PyFREQ for your xTB hessians
04 Apr 2021
Konnichiwa minasan! Salut tout le monde! Olá pessoal! Hi everybody! Well, today is a very special day for those who observe Easter. So, to those of you, I hope you all have a nice and wonderful Easter full of its spiritual meaning. I’m not catholic, but I know resurrection plays a deep role in the catholic heart, and quite frankly speaking, this is what we all need today: to resurrect from our death. You know, this COVID-19 thing put us all down, and we need to find the strength to carry on.
Well, to take a ride on that thought of carrying on I came here to tell you all what I’ve been doing those days. Actually, I got to work with GFN-xTB since it was made public, and I’m a big fan of it. To those who may not know, GFN-xTB is a great tight-binding method with Grimme’s parameters for almost the entire periodic table. A more in-depth presentation of it can be found in Grimme’s paper here, which is pretty easy to follow along.
So, as you might know, I’m also a fan of Python and PyMOL - why not? - and I like to work with Python as much as I can. As a computational chemist, one of our duties when working with molecules is making sure they are in a local minimum, i.e., after relaxing their structures one must make sure that structure is within its energy minimum, and is not going to jump to another state. For that, guess: we compute hessians. Well, actually that is a very simple concept that we use to testify if the second derivatives with respect to the energy do return you a positive value. Called eigenvalue or normal mode, a molecule will be said to be in a local minimum if all returned eigenvalues from its hessian computation are positive.
Well, actually I think most of you are also computational chemists or are studying it somehow. Therefore, as we are used to saying here in Brazil: “I’m not going to teach the priest how to do the mass”. On contrary, what I want to do is just to present this pretty tool I made using Python whose result can be assessed by PyMOL - or any other given tool that is capable of rendering xyz files such as Avogadro, Chimera, GausView, etc.
Here is the deal, suppose you have the structure shown in figure 1 to be optimized under GFN-xTB. That is a beta-cyclodextrin structure and has a total of 217 atoms. To perform such relaxation under the DFT level from a poor structural guess would be a waste of time. Therefore, it might be good to let GFN-xTB do this first step prior to a more advanced computational step.
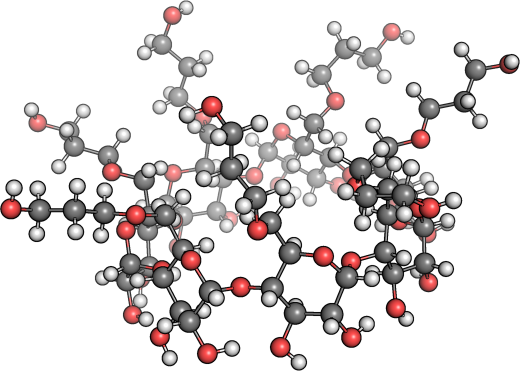 Figure 1: Just a shy beta-cyclodextrin molecular structure.
Figure 1: Just a shy beta-cyclodextrin molecular structure.
Well, supossing you have labelled your structure as bcd.xyz you may submit it to GFN-xTB like the example below:
xtb --opt tight --gfn 2 --acc .0001 --grad bcd.xyz > bcd.outPlease, pay atention that this is just an example and that’s not the right thing to do for all cases, and even for this one you might use a better approach. I’m just showing an example here, ok? So, after the convergence of this job you will end up with a file named xtbopt.xyz, which is the optimized geometry you want to assess the hessian. So, you will submit it to your desired hessian calculation as the example below:
xtb --hess --gfn 2 --acc .0001 --grad xtbopt.xyz > bcd.hess.outNow, if everything run just fine you might have got beyond other files two particular ones called g98.out and vibspectrum. Those files will be used by us to extract the normal modes of our system in order to visualize an animation of their behaviour, cool eh? Brace yourself, because here it goes:
#!/usr/bin/env python
# coding: utf-8
from sys import argv
from getopt import getopt, GetoptError
from numpy import linspace, array, append, loadtxt
from concurrent.futures import ProcessPoolExecutor as PPE
from functools import partial
from os.path import isfile
def PT(Z):
return {'1':'H','2':'He', '3':'Li', '4':'Be', '5':'B', '6':'C', '7':'N',\
'8':'O', '9':'F', '10':'Ne', '11':'Na', '12':'Mg', '13':'Al',\
'14':'Si', '15':'P', '16':'S', '17':'Cl', '18':'Ar', '19':'K',\
'20':'Ca', '21':'Sc', '22':'Ti', '23':'V', '24':'Cr', '25':'Mn',\
'26':'Fe', '27':'Co', '28':'Ni', '29':'Cu', '30':'Zn', '31':'Ga',\
'32':'Ge', '33':'As', '34':'Se', '35':'Br', '36':'Kr', '37':'Rb',\
'38':'Sr', '39':'Y', '40':'Zr', '41':'Nb', '42':'Mo', '43':'Tc',\
'44':'Ru', '45':'Rh', '46':'Pd', '47':'Ag', '48':'Cd', '49':'In',\
'50':'Sn', '51':'Sb', '52':'Te', '53':'I', '54':'Xe', '55':'Cs',\
'56':'Ba', '57':'La', '58':'Ce', '59':'Pr', '60':'Nd', '61':'Pm',\
'62':'Sm', '63':'Eu', '64':'Gd', '65':'Tb', '66':'Dy', '67':'Ho',\
'68':'Er', '69':'Tm', '70':'Yb', '71':'Lu', '72':'Hf', '73':'Ta',\
'74':'W', '75':'Re', '76':'Os', '77':'Ir', '78':'Pt', '79':'Au',\
'80':'Hg', '81':'Tl', '82':'Pb', '83':'Bi', '84':'Po', '85':'At',\
'86':'Rn', '87':'Fr', '88':'Ra', '89':'Ac', '90':'Th', '91':'Pa',\
'92':'U', '93':'Np', '94':'Pu', '95':'Am', '96':'Cm', '97':'Bk',\
'98':'Cf', '99':'Es', '100':'Fm', '101':'Md', '102':'No',\
'103':'Lr', '104':'Rf', '105':'Db', '106':'Sg', '107':'Bh',\
'108':'Hs', '109':'Mt', '110':'Ds', '111':'Rg', '112':'Cn',\
'113':'Nh', '114':'Fl', '115':'Mc', '116':'Lv', '117':'Ts',\
'118':'Og'}[Z]
def hesscheck(hess):
if not isfile(hess):
print('\n%s file not found.\n' % hess)
exit(1)
else: pass
def switcher(nfreq):
cond = nfreq % 3
if cond == 1: return 2
elif cond == 2: return 5
elif cond == 0: return 8
def interp(x, y, z, a, n, w):
for i, xs in enumerate(x):
w.write('{:d}\n\n'.format(n))
for j, XS in enumerate(xs):
w.write('{:s} {:8.4f} {:8.4f} {:8.4f}\n'.format(PT(a[j]), XS,\
y[i][j], z[i][j]))
def parsehess(nfreq, hess):
if nfreq == 'all':
modes, ir = loadtxt(open('vibspectrum').readlines()[:-1],\
skiprows = 3, usecols = [0, 4],\
unpack = True, dtype = [('mode', 'int'),\
('IR', '<U16')])
modes = array([modes[i] for i, c in enumerate(ir) if c == 'YES'])
nmodes = array([modes[i] for i, c in enumerate(ir) if c == '-'])
modes -= len(nmodes)
partials = partial(getfreq, hess)
with PPE() as executor:
results = executor.map(partials, modes)
else:
getfreq(hess, int(nfreq))
def getfreq(hess, nfreq):
ac, locfreq = False, False
natom = 0
atms = array([])
X, Y, Z = array([]), array([]), array([])
fx, fy, fz = array([]), array([]), array([])
with open(hess, 'r') as tf:
for line in tf:
ls = line.split()
if 'Center' in ls:
next(tf)
next(tf)
ac = True
elif len(ls) == 1: ac = False
elif len(ls) == 3:
try:
for mode in ls:
if nfreq == int(mode): locfreq = True
except: pass
elif ac:
CN, AN, AT, x, y, z = ls
atms = append(atms, AN)
X = append(X, float(x))
Y = append(Y, float(y))
Z = append(Z, float(z))
natom += 1
elif 'Atom' in ls and 'AN' in ls and locfreq:
lfreq = switcher(nfreq)
for j in range(natom):
ln = next(tf).split()
fx = append(fx, float(ln[lfreq]))
fy = append(fy, float(ln[lfreq+1]))
fz = append(fz, float(ln[lfreq+2]))
locfreq = False
minx, maxx = X - fx, X + fx
miny, maxy = Y - fy, Y + fy
minz, maxz = Z - fz, Z + fz
interfx = linspace(minx, maxx, 10)
interfy = linspace(miny, maxy, 10)
interfz = linspace(minz, maxz, 10)
interbx = linspace(maxx, minx, 10)
interby = linspace(maxy, miny, 10)
interbz = linspace(maxz, minz, 10)
with open('freq.%05d.xyz' % int(nfreq), 'w') as wfreq:
interp(interfx, interfy, interfz, atms, natom, wfreq)
interp(interbx, interby, interbz, atms, natom, wfreq)
def usage():
print('''
Hi there, here is your tip for calling PyFREQ.py:
PyFREQ.py --mode <int|all>
NOTA BENE:
1) The only option you can give to mode is an integer number
or 'all', which shouldn't be in quotation marks;
2) PyFREQ.py assumes g98.out and vibspectrum are within its
working directory.
''')
exit(0)
if __name__ == '__main__':
try:
opts, args = getopt(argv[1:], 'hm:', ['help', 'mode='])
for o, a in opts:
if o in ['-h', '--help']:
usage()
elif o in ['-m', '--mode']:
nfreq = a
else:
print('\n\nUnhandled option.\n\n')
exit(0)
except GetoptError as err:
print('\n', str(err))
hesscheck('g98.out')
hesscheck('vibspectrum')
try:
if nfreq == 'all' or int(nfreq): pass
except:
usage()
parsehess(nfreq, 'g98.out')Now, you may copy it to a file named PyFREQ.py. Once that is done you might test it out like this:
chmod +x PyFREQ.py
./PyFREQ.py --helpFinally, you can select what you want to do. Would you like to get all normal modes at once, or would you prefer to retrieve only a given value? For instance, suppose you want to see the first normal mode. That would be something like this:
./PyFREQ.py --mode 1For me, that gave me back a file labelled as freq.00001.xyz. Notice that all files will be given as freq.N.xyz, in which N is the normal mode number with at least 5 zeros as padding. Well, why 5 zeros as padding? I don’t know, my past me thought it would be enough. But you may change it easily by setting a different value in the code. :)
Well, to open up and see this normal mode I would recommend using PyMOL as this:
pymol freq.00001.xyz -d 'show spheres; set sphere_scale, .25; show lines; hide sticks; color gray, symbol C'Which for me gave me the result seen in figure 2.
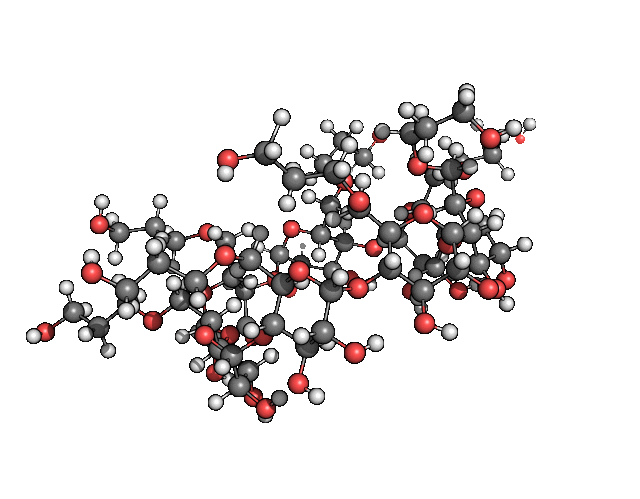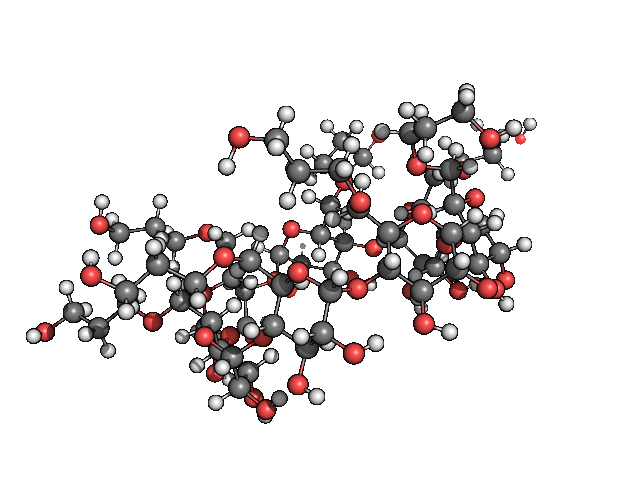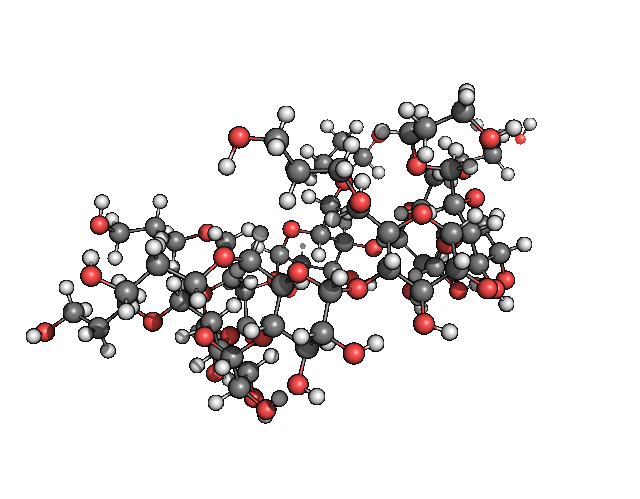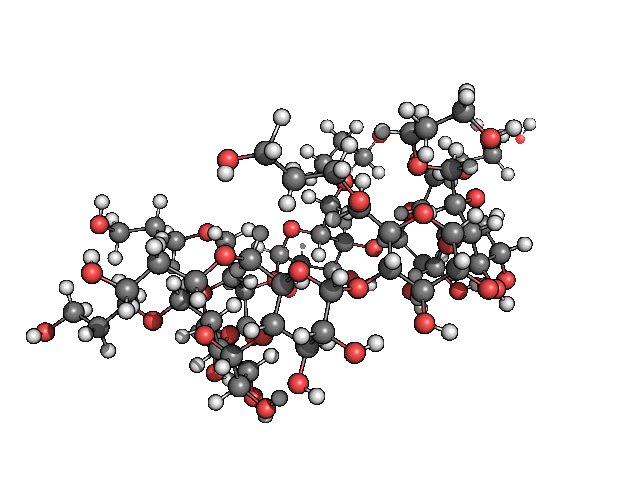 Figure 2: The cutest normal mode you have ever laid your eyes on.
Figure 2: The cutest normal mode you have ever laid your eyes on.
That’s it, folks. I hope you like it as much as I do. :)
Take care.
Byeeeeeee.